We will here use the create_table_forestplot and
dessin_forest_plot functions in the Data & Functions page.
Download Forest_autom.R
# Libraries
library(rms) # or library(survival)
library(cmprsk)
library(forestplot) # version 3.1.1
# Creation of dataset
set.seed(143)
lung$bras <- sample(c('A','B'), nrow(lung), replace=TRUE)
lung$status2 <- sample(0:2, nrow(lung), replace=TRUE, prob=c(0.6, 0.25, 0.15))
lung$status <- lung$status-1
lung$ecog[lung$ph.ecog %in% c(0,1)] <- '01'
lung$ecog[lung$ph.ecog %in% c(2,3)] <- '23'
lung <- lung[,c("bras","time","status","status2","age","sex","ecog","meal.cal")]create_table_forestplot function
cov <- c("age","sex","ecog","meal.cal")
quali <- rep(1,length(cov))
quali[cov %in% c("age","meal.cal")] <-0
tabforest1 <- create_table_forestplot(data_frame=lung,
covariables=cov,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras",
var_dinteret="status",
delai_dinteret="time")| Subset | A (Events/Pts) | B (Events/Pts) | HR (95%CI) | Pvalue_interaction |
|---|---|---|---|---|
| age | > 0.2 | |||
| ≤63 | 46/67 | 34/50 | 1.17 (0.75 - 1.82) | |
| >63 | 51/63 | 34/48 | 0.97 (0.63 - 1.5) | |
| sex | > 0.2 | |||
| 1 | 62/77 | 50/61 | 1.09 (0.75 - 1.58) | |
| 2 | 35/53 | 18/37 | 0.91 (0.51 - 1.61) | |
| ecog | > 0.1 | |||
| 01 | 67/98 | 52/78 | 1.22 (0.85 - 1.75) | |
| 23 | 30/32 | 15/19 | 0.71 (0.38 - 1.32) | |
| meal.cal | 0.99 | |||
| ≤975 | 40/55 | 25/36 | 1.1 (0.67 - 1.82) | |
| >975 | 37/48 | 32/42 | 1.1 (0.68 - 1.76) | |
| All patients | 97/130 | 68/98 | 1.06 (0.78 - 1.45) |
Here is another example where optional parameters are used:
nom.var <- c("Age (years)","Gender","ECOG","Calories consumed")
levels.var <- c("Male","Female","0-1","2-3")
tabforest2 <- create_table_forestplot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status",
delai_dinteret="time",
percent=TRUE)| Subset | Arm A (Events/Pts) | Arm B (Events/Pts) | HR (95%CI) | Pvalue_interaction |
|---|---|---|---|---|
| Age (years) | > 0.2 | |||
| ≤63 | 46/67 (69%) | 34/50 (68%) | 1.17 (0.75 - 1.82) | |
| >63 | 51/63 (81%) | 34/48 (71%) | 0.97 (0.63 - 1.5) | |
| Gender | > 0.2 | |||
| Male | 62/77 (81%) | 50/61 (82%) | 1.09 (0.75 - 1.58) | |
| Female | 35/53 (66%) | 18/37 (49%) | 0.91 (0.51 - 1.61) | |
| ECOG | > 0.1 | |||
| 0-1 | 67/98 (68%) | 52/78 (67%) | 1.22 (0.85 - 1.75) | |
| 2-3 | 30/32 (94%) | 15/19 (79%) | 0.71 (0.38 - 1.32) | |
| Calories consumed | 0.99 | |||
| ≤975 | 40/55 (73%) | 25/36 (69%) | 1.1 (0.67 - 1.82) | |
| >975 | 37/48 (77%) | 32/42 (76%) | 1.1 (0.68 - 1.76) | |
| All patients | 97/130 (75%) | 68/98 (69%) | 1.06 (0.78 - 1.45) |
In the example above, the covariates, the levels of the categorical variables and the treatment arms are named. The percentages of the numbers are also displayed.
dessin_forest_plot function
Here is an example of a forest plot without the optional arguments:
dessin_forest_plot(data_frame=lung,
covariables=cov,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras",
var_dinteret="status",
delai_dinteret="time")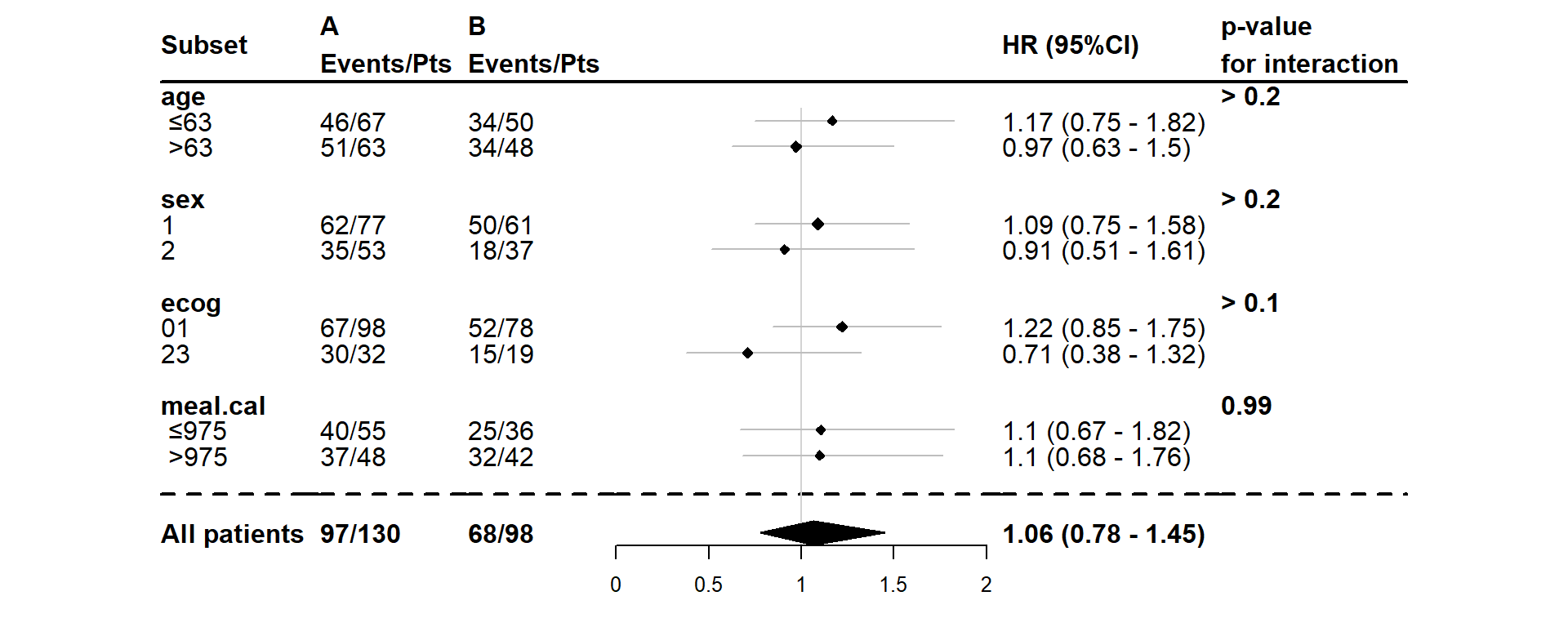
We can:
- Give a name to the covariates, categorical variable levels and treatment arms
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status",
delai_dinteret="time")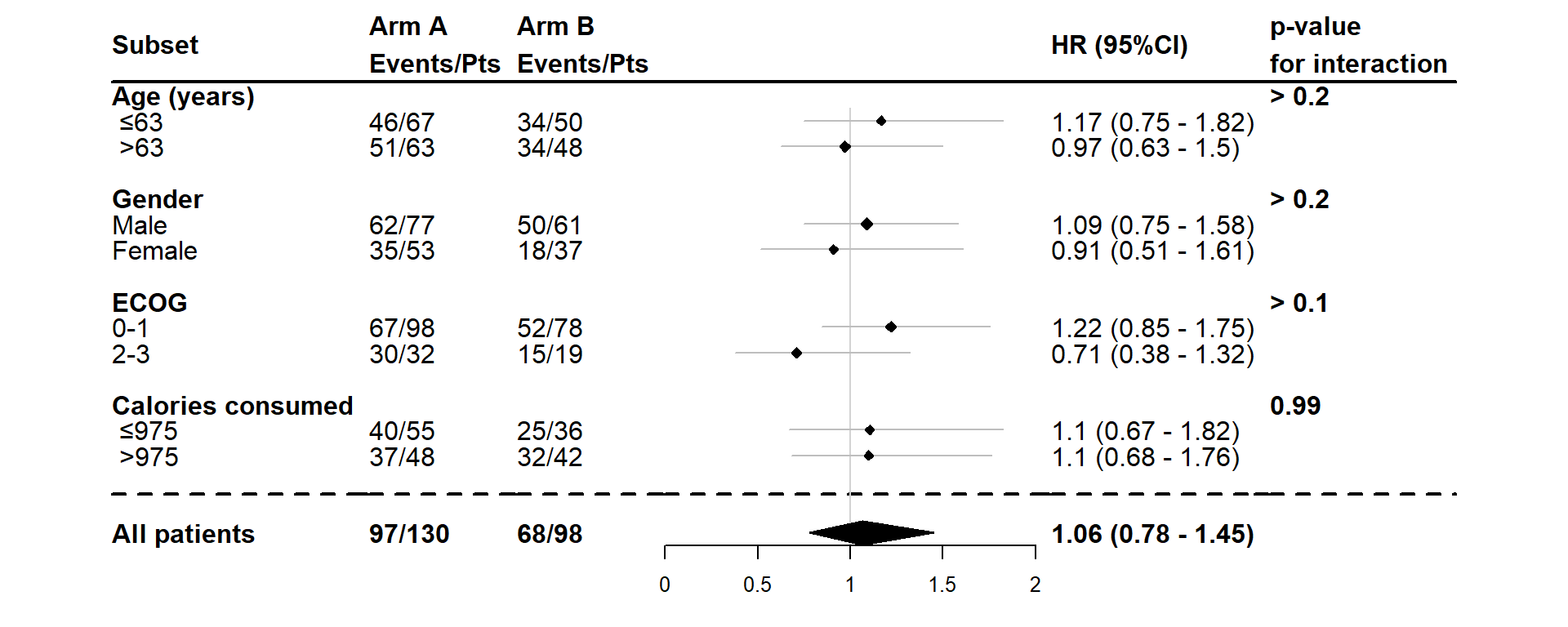
- Change the position and color of the graph, as well as the shape of the estimate
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status",
delai_dinteret="time",
pos_graphique=2,
col=fpColors(box="palegreen4", lines="palegreen3", summary="palegreen4"),
forme=fpDrawNormalCI)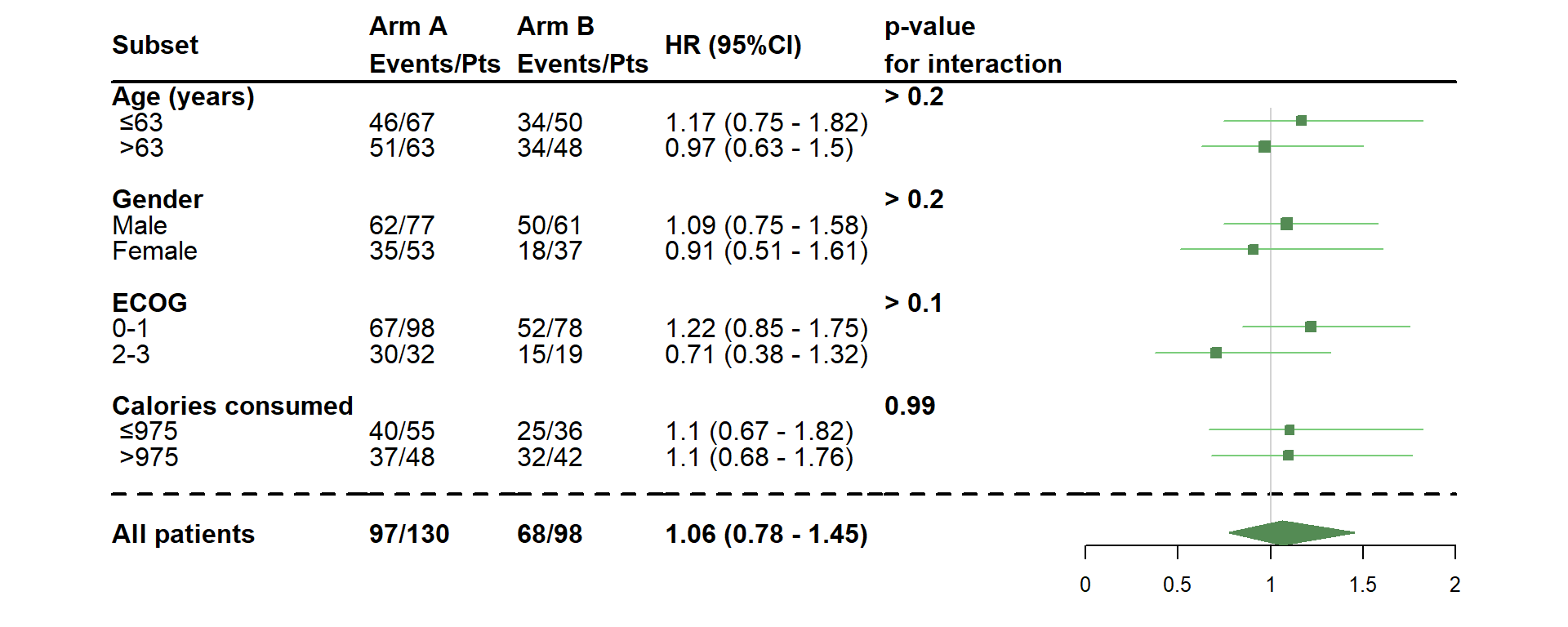
- Change the size of the estimates
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status",
delai_dinteret="time",
pos_graphique=2,
col=fpColors(box="palegreen4", lines="palegreen3", summary="palegreen4"),
forme=fpDrawNormalCI,
boxsize=0.8)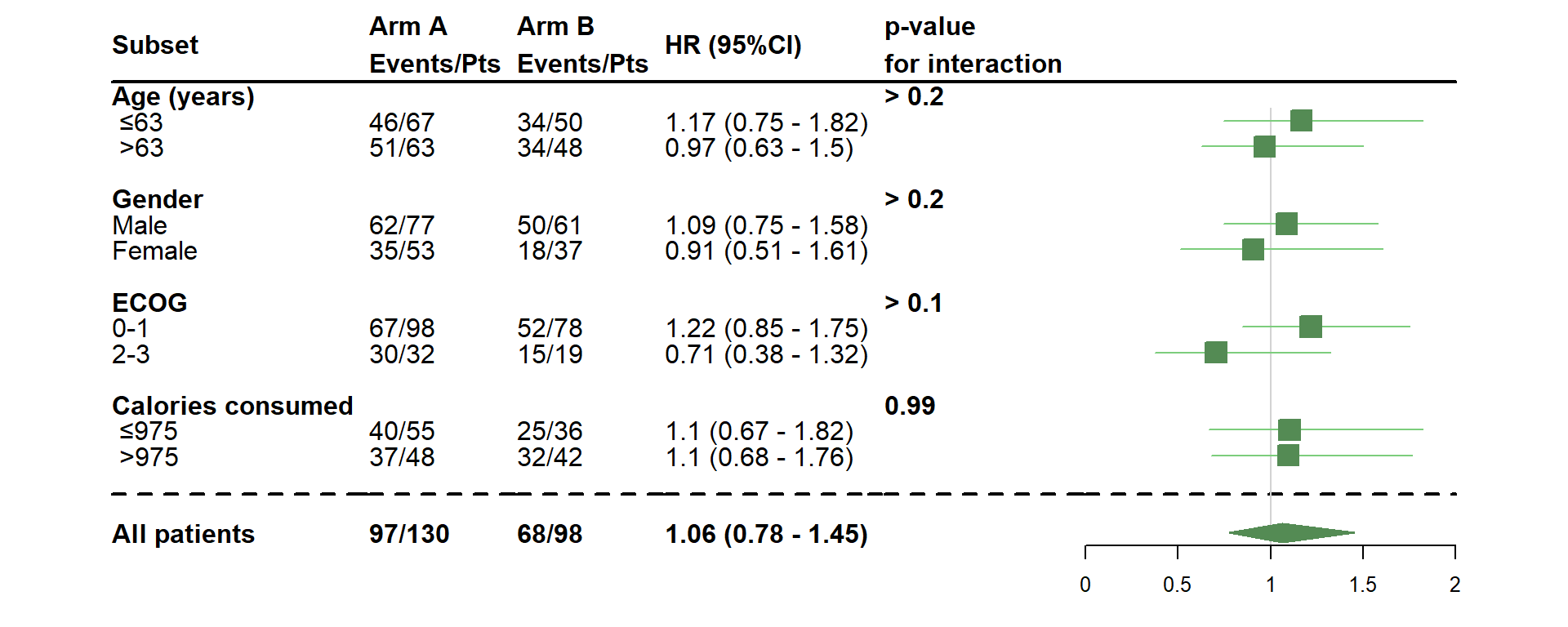
- Change the threshold values of quantitative variables
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status",
delai_dinteret="time",
cut_off=c(70,950),
pos_graphique=2,
col=fpColors(box="palegreen4", lines="palegreen3", summary="palegreen4"),
forme=fpDrawNormalCI,
boxsize=0.8)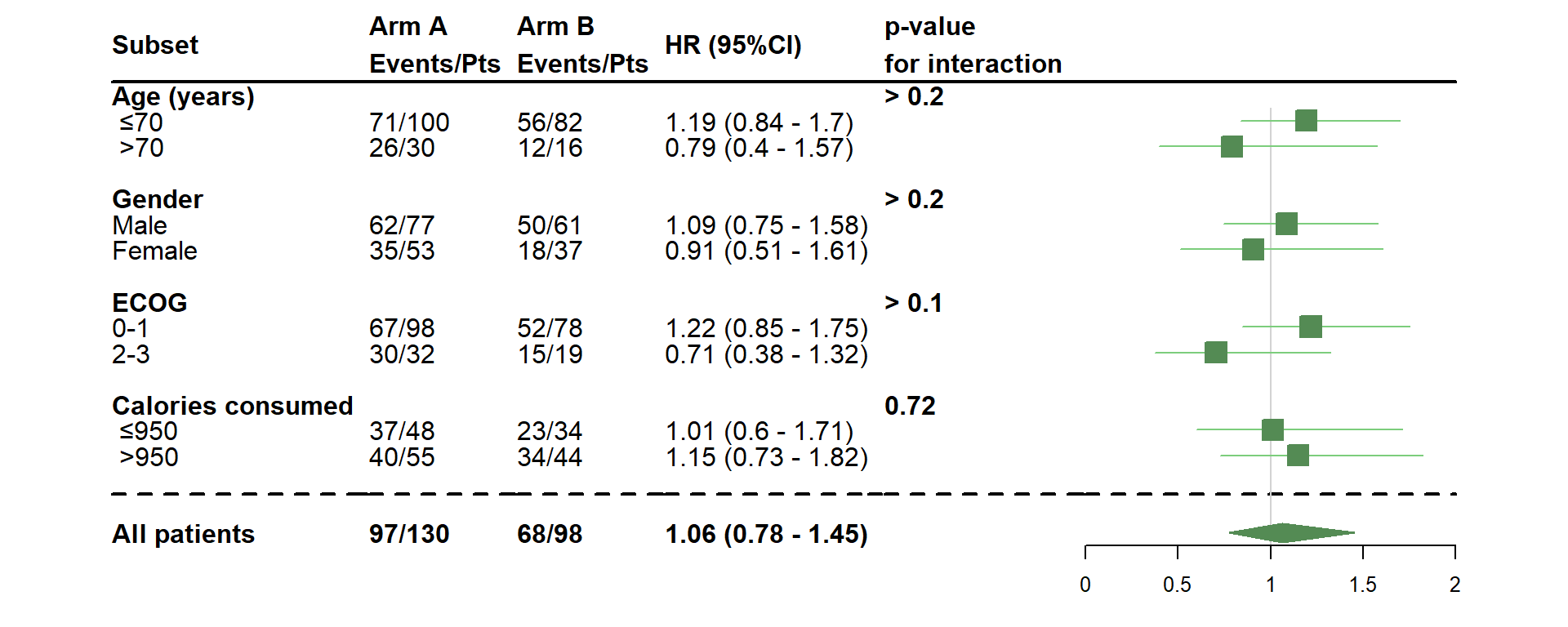
- Choose a log scale
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status",
delai_dinteret="time",
cut_off=c(70,950),
xlog=TRUE,
pos_graphique=2,
col=fpColors(box="palegreen4", lines="palegreen3", summary="palegreen4"),
forme=fpDrawNormalCI)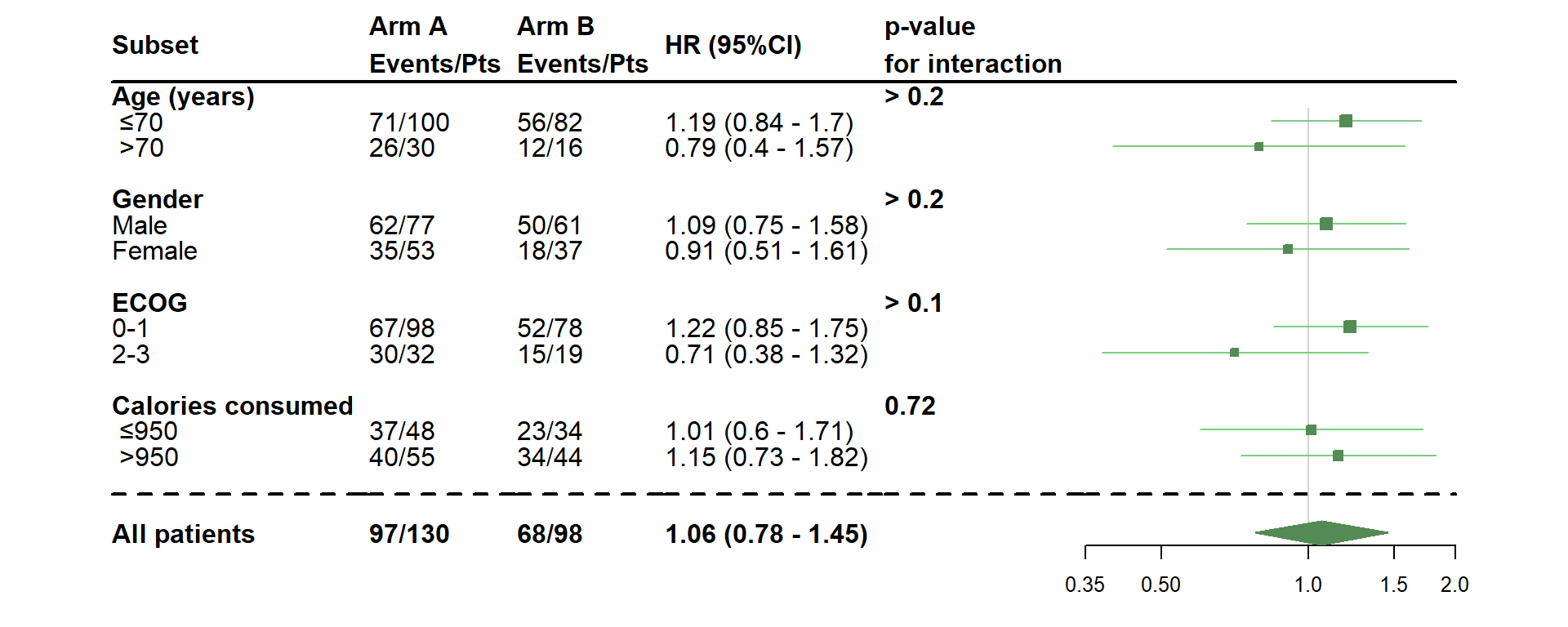
- Specified the difference in direction below the axis
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status",
delai_dinteret="time",
cut_off=c(70,950),
xlog=TRUE,
pos_graphique=2,
col=fpColors(box="palegreen4", lines="palegreen3", summary="palegreen4"),
forme=fpDrawNormalCI,
favors=c("Favors A","Favors B"),
favors_fontsize=10)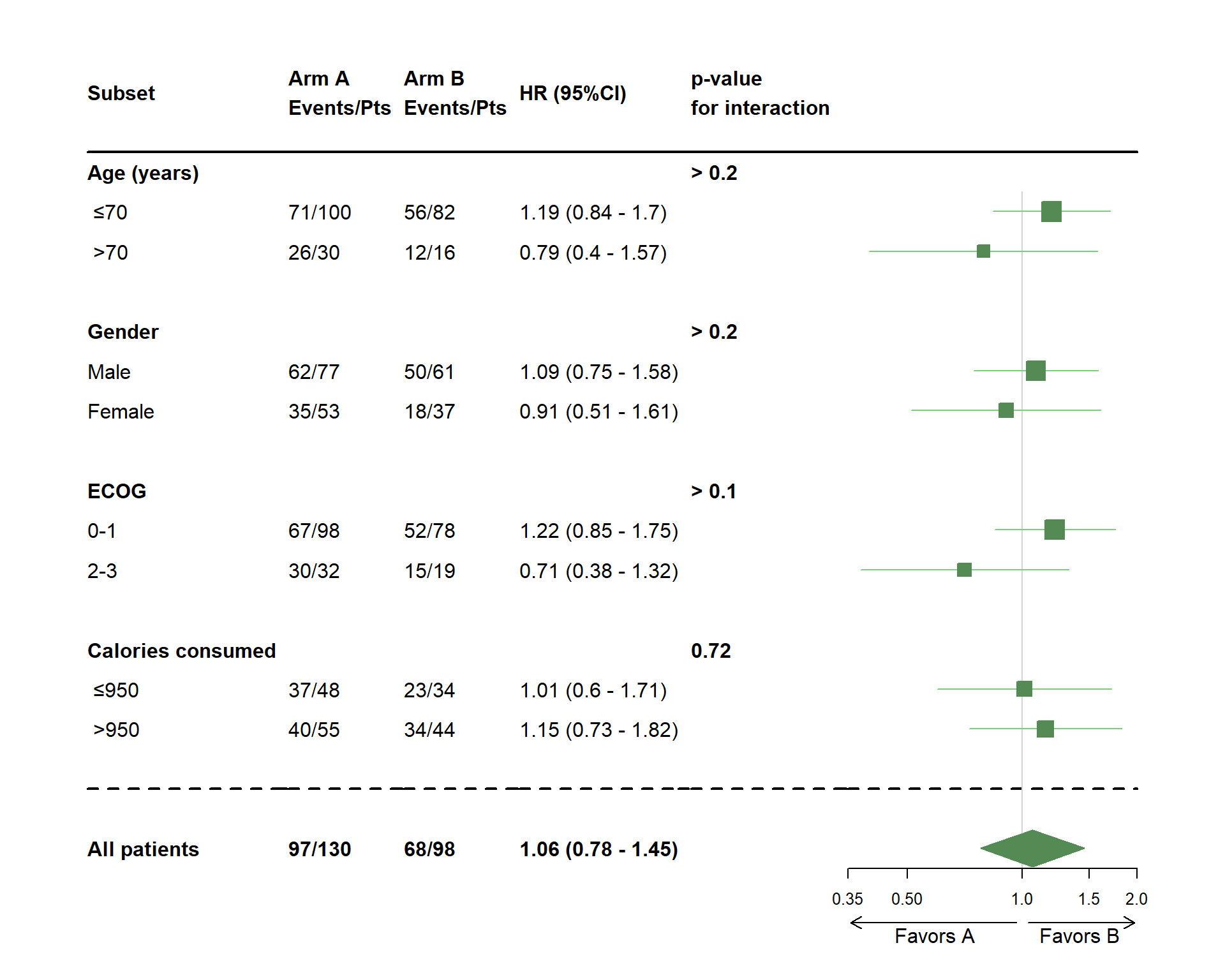
- Add medians
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="cox",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status",
delai_dinteret="time",
cut_off=c(70,950),
median=TRUE,
xlog=TRUE,
pos_graphique=2,
col=fpColors(box="palegreen4", lines="palegreen3", summary="palegreen4"),
forme=fpDrawNormalCI,
favors=c("Favors A","Favors B"),
favors_fontsize=10)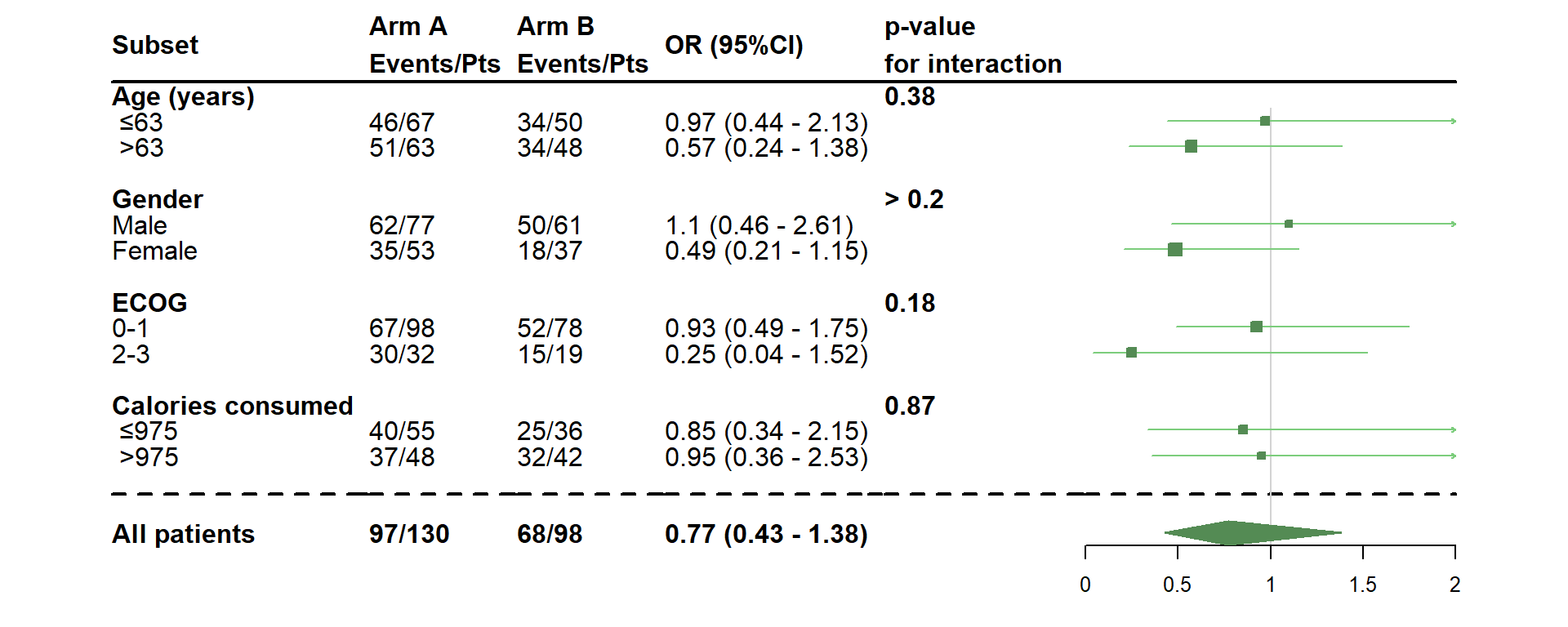
- Use a logistic model
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="log",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status",
pos_graphique=2,
col=fpColors(box="palegreen4", lines="palegreen3", summary="palegreen4"),
forme=fpDrawNormalCI)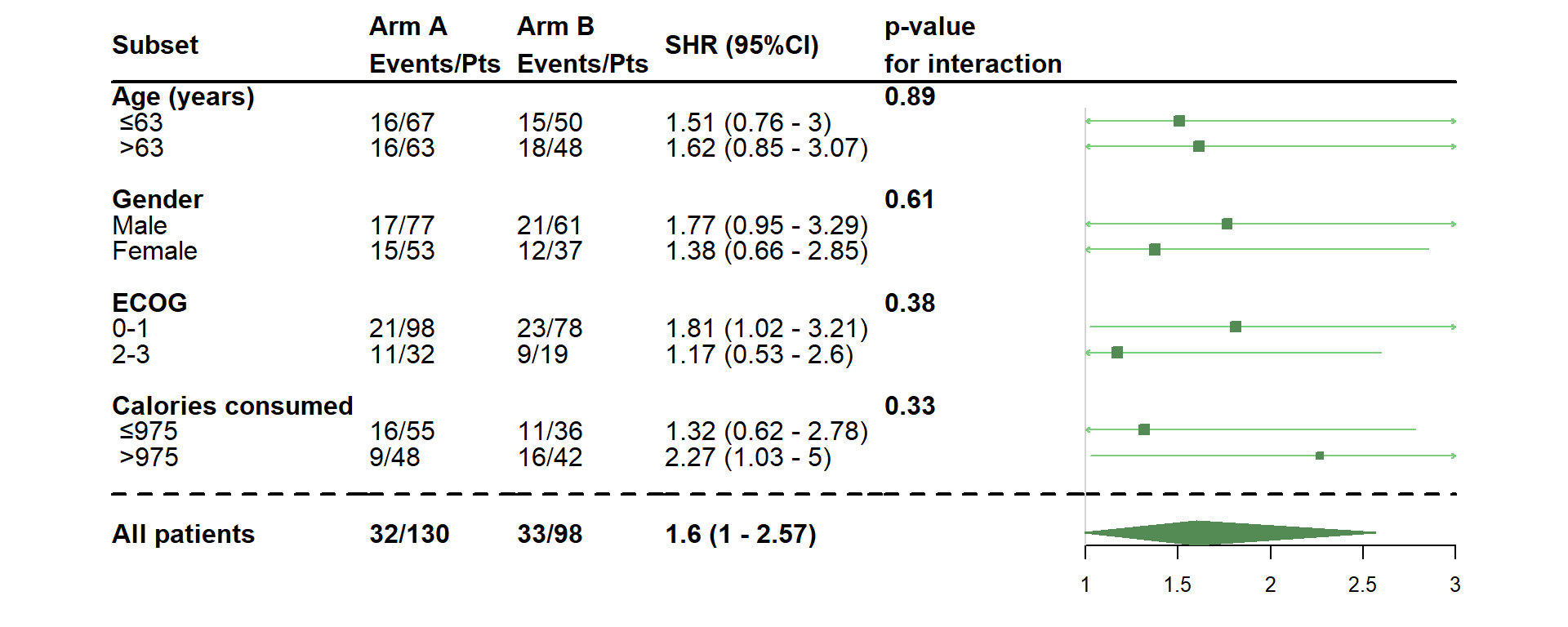
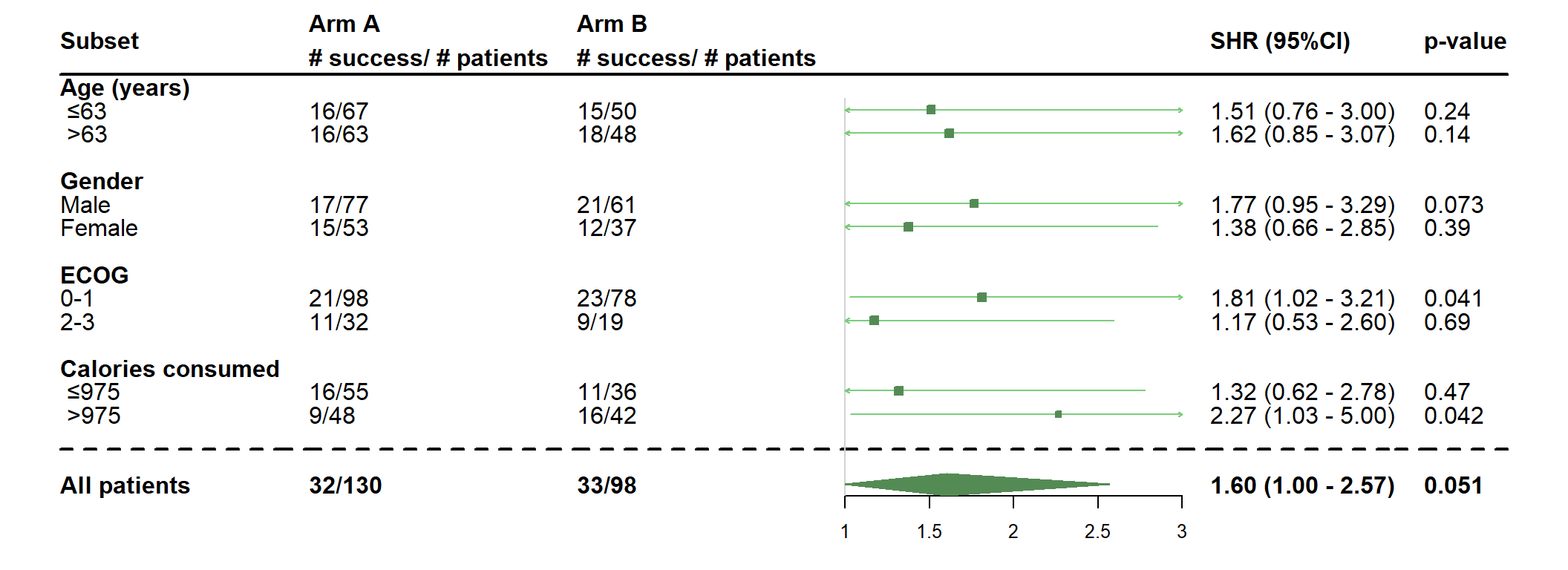
- Use a Fine & Gray model
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="finegray",
bras="bras", nom_bras=c("Arm A","Arm B"),
var_dinteret="status2", failcode=1,
delai_dinteret="time",
pos_graphique=2,
col=fpColors(box="palegreen4", lines="palegreen3", summary="palegreen4"),
forme=fpDrawNormalCI)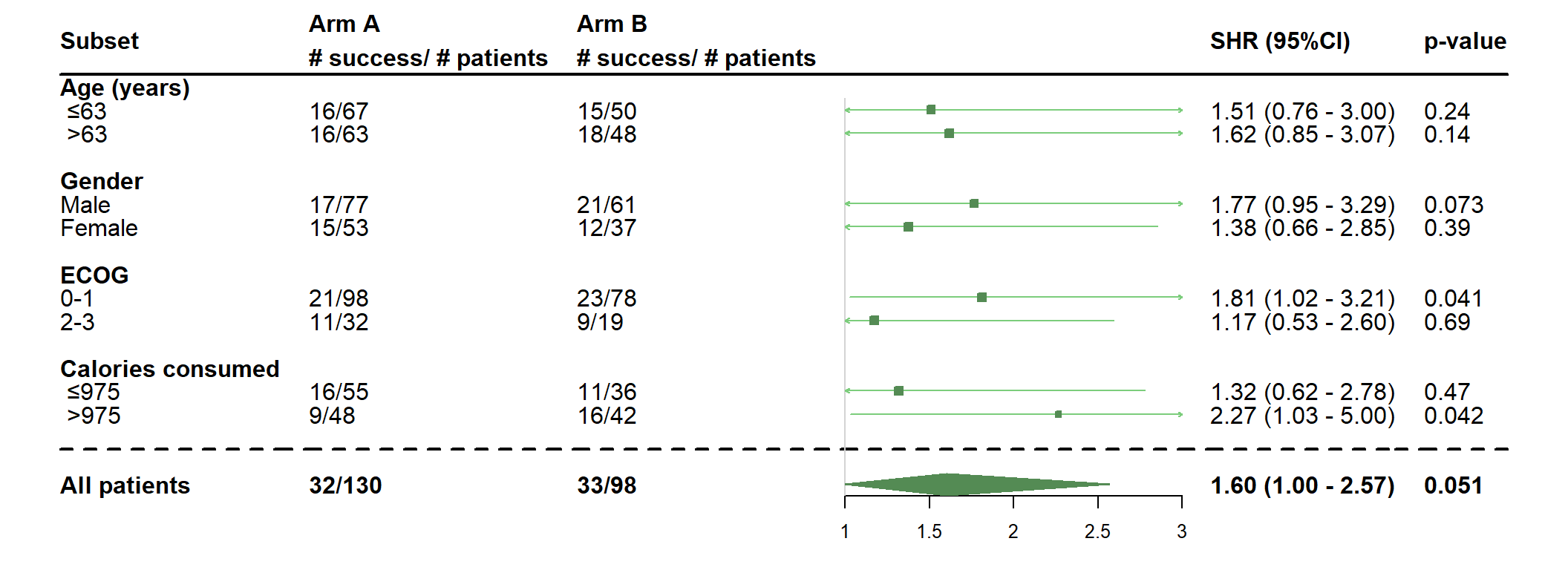
- Use more options : remove interaction p-values, add p-values for each subgroup, change the place of the plot and fix the number of digits
dessin_forest_plot(data_frame=lung,
covariables=cov, nom_var=nom.var,
levels_var=levels.var,
var_quali_ou_quanti=quali,
modele_souhaite="finegray",
bras="bras", nom_bras=c("Arm A","Arm B"), label_bras="# success/ # patients",
var_dinteret="status2", failcode=1,
delai_dinteret="time",
digit=2, fixed_digit=TRUE,
test=NULL,
pos_graphique=0,
col=fpColors(box="palegreen4", lines="palegreen3", summary="palegreen4"),
forme=fpDrawNormalCI,
print_pval = TRUE)