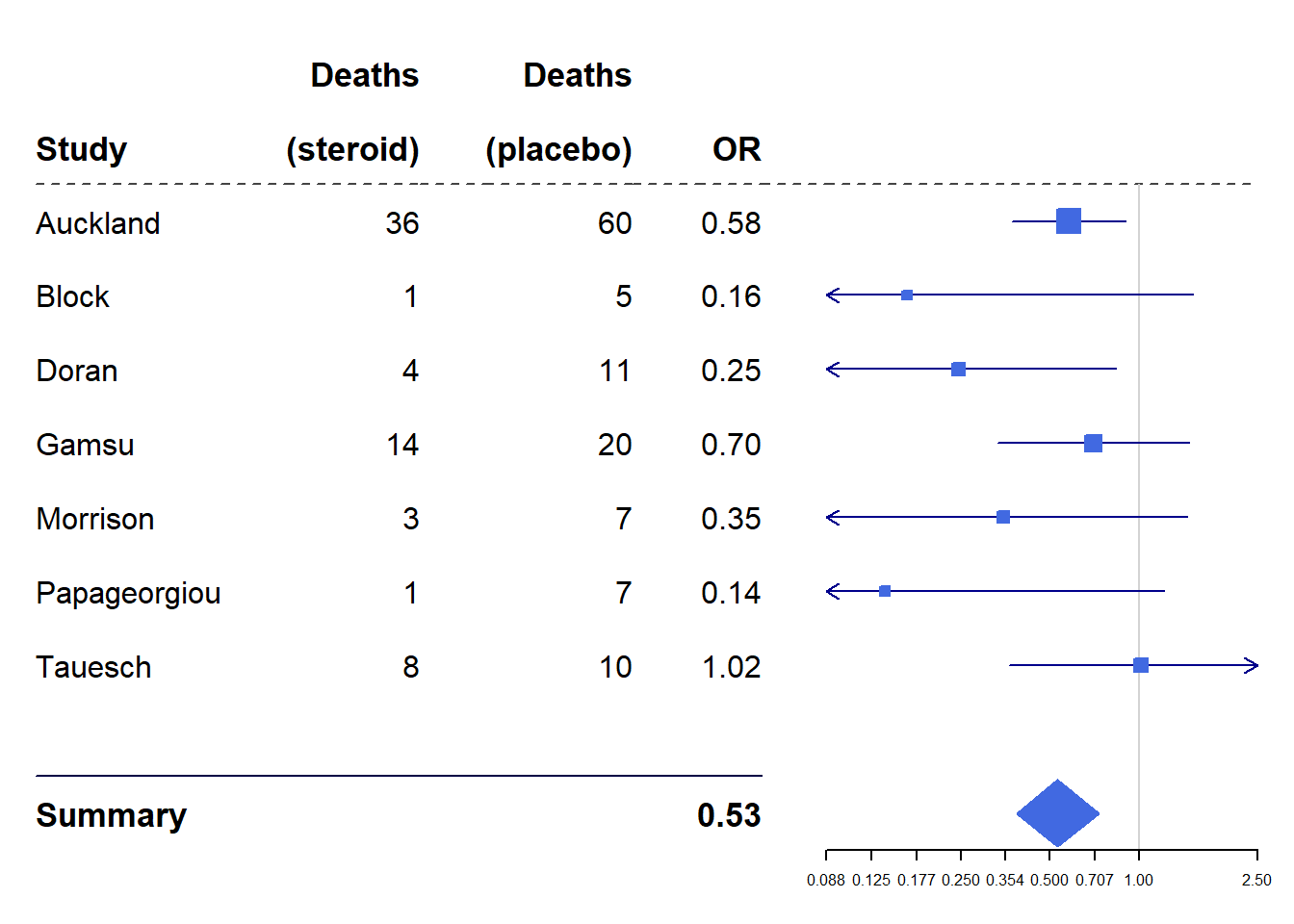
Tips to read
- each line corresponds to a study
- the symbol (here square or diamond) corresponds to the estimate of the parameter studied (here a relative risk)
- the size of the symbol is proportional to the precision
- the vertical bar is the value of the parameter in the absence of effect (here 1 because it is a relative risk)
- the horizontal bars correspond to the confidence interval of the reported parameter (here the relative risk)
- if the symbol is on the right of the vertical line, it means that the link between the treatment (here inspiratory muscle training) and the endpoint (here survival) is positive
- if the symbol is to the left of the vertical line, it means that the link between the treatment (here inspiratory muscle training) and the endpoint (here survival) is negative
- if the horizontal bar crosses the vertical bar then the link is not significant
- the diamond represents the parameter estimated from all the studies
Meta-analysis forest plot
# Libraries
library(forestplot)
library(tibble)
library(tidyjson)
library(dplyr)
# Cochrane data
base_data <- tibble(mean = c(0.578, 0.165, 0.246, 0.700, 0.348, 0.139, 1.017),
lower = c(0.372, 0.018, 0.072, 0.333, 0.083, 0.016, 0.365),
upper = c(0.898, 1.517, 0.833, 1.474, 1.455, 1.209, 2.831),
study = c("Auckland", "Block", "Doran", "Gamsu", "Morrison", "Papageorgiou", "Tauesch"),
deaths_steroid = c("36", "1", "4", "14", "3", "1", "8"),
deaths_placebo = c("60", "5", "11", "20", "7", "7", "10"),
OR = c("0.58", "0.16", "0.25", "0.70", "0.35", "0.14", "1.02"))
summary <- tibble(mean = 0.531,
lower = 0.386,
upper = 0.731,
study = "Summary",
OR = "0.53",
summary = TRUE)
header <- tibble(study = c("", "Study"),
deaths_steroid = c("Deaths", "(steroid)"),
deaths_placebo = c("Deaths", "(placebo)"),
OR = c("", "OR"),
summary = TRUE)
empty_row <- tibble(mean = NA_real_)
cochrane_output_df <- bind_rows(header,
base_data,
empty_row,
summary)
cochrane_output_df %>%
forestplot(labeltext = c(study, deaths_steroid, deaths_placebo, OR),
is.summary = summary,
clip = c(0.1, 2.5),
hrzl_lines = list("3" = gpar(lty = 2),
"11" = gpar(lwd = 1, columns = 1:4, col = "#000044")),
xlog = TRUE,
col = fpColors(box = "royalblue",
line = "darkblue",
summary = "royalblue",
hrz_lines = "#444444"))