Data
We load the packages we will use:
We use the data frame called aSAH from the package
pROC:
# Data
data(aSAH)
aSAH1 <- aSAH[,c("outcome","s100b")]
rocobj <- roc(aSAH1$outcome, aSAH1$s100b, ci=T)aSAH format:
A data frame containing 113 observations with 2 variables we will
use:
outcome MCC type
s100b
Concentration of S100B
| outcome | s100b |
|---|---|
| Good | 0.13 |
| Good | 0.14 |
| Good | 0.10 |
| Good | 0.04 |
| Poor | 0.13 |
| Poor | 0.10 |
| Good | 0.47 |
| Poor | 0.16 |
ggroc function
ggroc(
data, a roc object from the roc
function, or a list of roc objects
aes, the name of
the aesthetics for geom_line to map to the different ROC curves
supplied. Use “group” if you want the curves to appear with the same
aestetic, for instance if you are faceting instead
legacy.axes, a logical indicating if the specificity axis
(x axis) must be plotted as as decreasing “specificity” (FALSE, the
default) or increasing “1 - specificity” (TRUE) as in most legacy
software
...
)
ROC curve
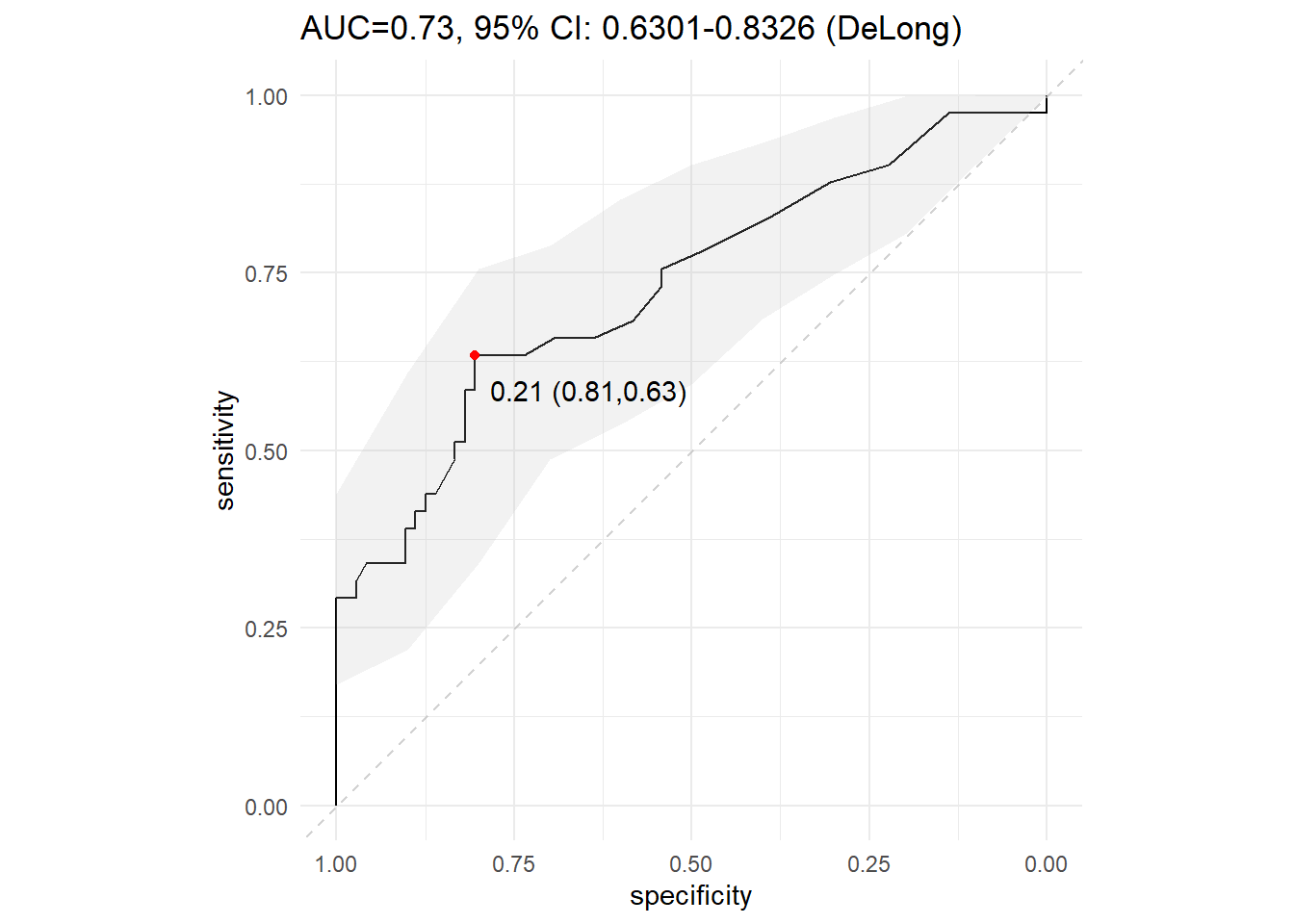
# Advanced ROC curve
ci.se.rocobj <- ci(rocobj, of="se", boot.n=2000) # confidence interval of the sensitivity
dat.ci <- data.frame(x = as.numeric(rownames(ci.se.rocobj)),
lower = ci.se.rocobj[, 1],
upper = ci.se.rocobj[, 3])
# selection of a threshold
dat.seuil=coords(rocobj, "best", best.method=c("youden"), ret=c("threshold", "sensitivity", "specificity"))
ggroc(rocobj) +
theme_minimal() +
geom_abline(slope=1, intercept = 1, linetype = "dashed", alpha=0.7, color = "grey") + # add abline and custom
coord_equal() + # ensures the units are equally scaled on the x-axis and on the y-axis
geom_ribbon(data = dat.ci, aes(x = x, ymin = lower, ymax = upper), fill = "grey", alpha= 0.2) + # generate confidence interval of sensibility
ggtitle(paste0("AUC=", round(rocobj$auc, 2), ", ", capture.output(rocobj$ci))) +
geom_point(data = tibble(se=dat.seuil$sensitivity, sp=dat.seuil$specificity), mapping = aes(x=sp, y=se), colour = "red") +
annotate("text", x=dat.seuil$specificity-0.16, y=dat.seuil$sensitivity-0.05,
label= paste0(round(dat.seuil$threshold, 2), " (", round(dat.seuil$specificity, 2), ",", round(dat.seuil$sensitivity, 2), ")"))