To reconstruct a Kaplan-Meier curve and generate pseudo individual
patient data (IPD), several steps are required. The example below is
based on the following image
(Download).
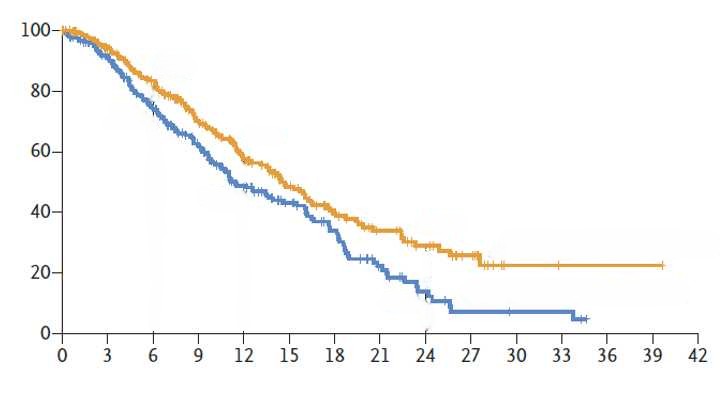
Step 1: Digitization
SurvdigitizeR is an R package that automates the
extraction of survival probabilities from Kaplan-Meier (KM) curves in
JPEG or PNG format. It simplifies the process of digitizing KM curves,
which is typically time-consuming and prone to error when done manually.
This package provides an efficient and accurate way to digitize KM
curves.
# Libraries
#devtools::install_github("Pechli-Lab/SurvdigitizeR")
library(SurvdigitizeR)
library(tidyverse)
library(ggplot2)
# Digitization
set.seed(1354534)
CourbSurv_time <- survival_digitize(img_path = "article_surv.jpg",
x_start = 0, x_end = 42, x_increment = 3,
y_start = 0, y_end = 1, y_increment = 0.2,
y_text_vertical = TRUE, # if the Y-axis labels are vertical or horizontal
censoring = TRUE, # if censoring is represented by black marks
line_censoring = TRUE, # if line censoring removal should be attempted
num_curves = 2)
CourbSurv_time %>%
ggplot(aes(x = time, y= St, color = as.factor(curve), group = curve)) +
geom_step() + ylim(0,1) + theme_bw() +
scale_x_continuous(breaks = seq(0,42,3)) +
scale_y_continuous(breaks = seq(0,1,0.2))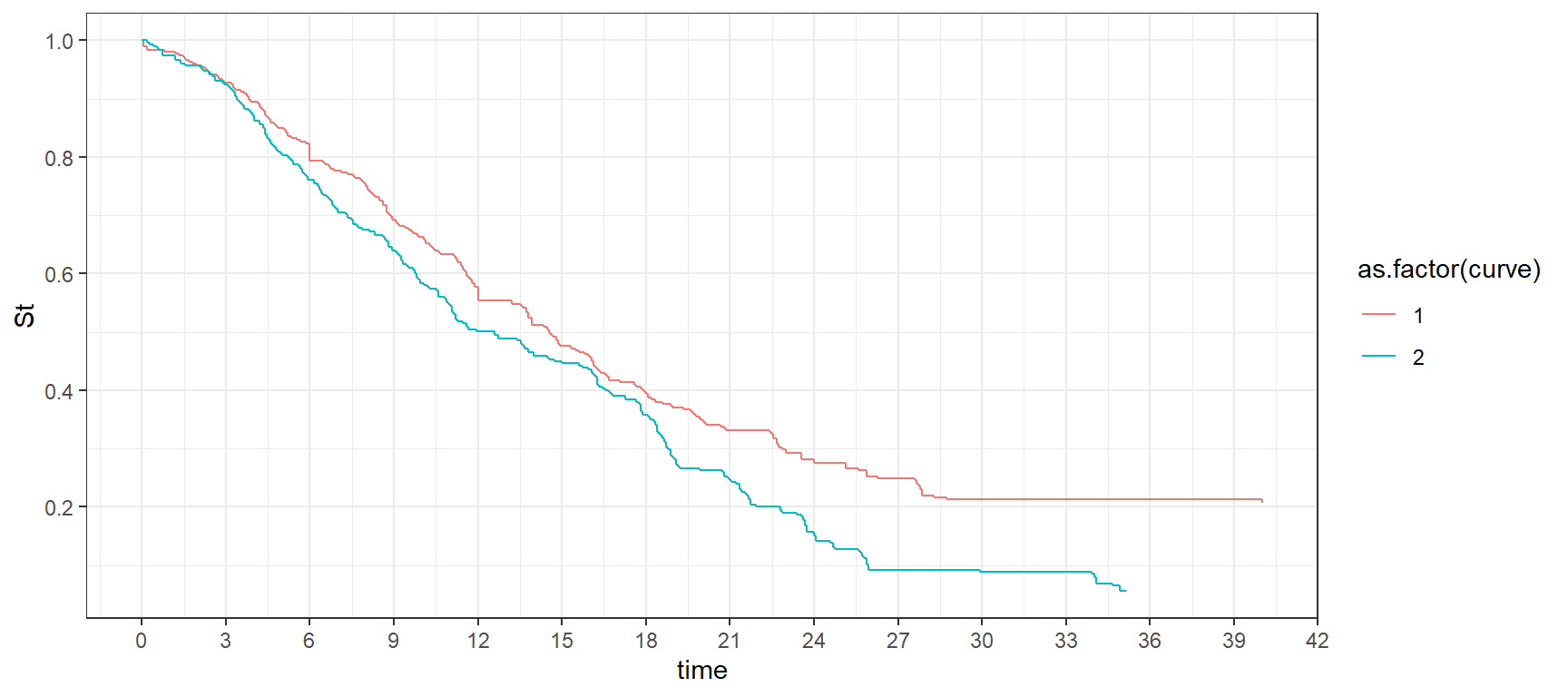
Step 2: Number at risk
In this step, the number of patients at risk at specific time points is reported. These values are used to align the digitized survival probabilities with the patient counts and improve reconstruction accuracy.
# Number at risk
N_timepoints <- 15
CourbSurv_nrisk <- data.frame(
Group = c(rep(1,N_timepoints),rep(2,N_timepoints)), # 2 groups
Interval = rep(1:N_timepoints,2),
Time = rep(c(0,3,6,9,12,15,18,21,24,27,30,33,36,39,42),2),
Lower = rep(NA,N_timepoints*2),
Upper = rep(NA,N_timepoints*2),
nrisk = c(246,219,185,134,94,65,45,30,21,12,2,1,1,1,0, # group 1
248,204,150,109,76,52,36,18, 9, 4,3,3,0,0,0) # group 2
)
# be careful to correctly identify which is group 1 and 2 in order to place them in the correct order (in "nrisk" argument)
for (g in unique(CourbSurv_nrisk$Group)) {
CourbSurv_nrisk$Lower[which(CourbSurv_nrisk$Group == g)] <-
sapply(CourbSurv_nrisk$Time[CourbSurv_nrisk$Group == g],
function(x){ min(CourbSurv_time$id[CourbSurv_time$curve==g &
CourbSurv_time$time>=x]) })
CourbSurv_nrisk$Upper[which(CourbSurv_nrisk$Group == g)] <-
c(sapply(CourbSurv_nrisk$Time[CourbSurv_nrisk$Group == g],
function(x){ max(CourbSurv_time$id[CourbSurv_time$curve==g &
CourbSurv_time$time<=x]) })[-1], max(CourbSurv_time$id[CourbSurv_time$curve==g]))
}
CourbSurv_nrisk <- CourbSurv_nrisk %>% filter(!is.infinite(Lower))
# Files .txt for each group
dir.create("digitization_txt")
write_tsv(CourbSurv_time %>% filter(curve==1),
"digitization_txt/CourbSurv1_time.txt")
write_tsv(CourbSurv_time %>% filter(curve==2),
"digitization_txt/CourbSurv2_time.txt")
write_tsv(CourbSurv_nrisk %>% filter(Group==1) %>% dplyr::select(-Group),
"digitization_txt/CourbSurv1_nrisk.txt")
write_tsv(CourbSurv_nrisk %>% filter(Group==2) %>% dplyr::select(-Group),
"digitization_txt/CourbSurv2_nrisk.txt")Step 3: Pseudo individual patient data
The digitized Kaplan Meier curves and the numbers at risk are
combined to reconstruct pseudo IPD with the function
digitise from the R package survHE.
# Libraries
library(survHE)
library(survminer)
library(readr)
# Pseudo IPD
digitise(surv_inp = "digitization_txt/CourbSurv1_time.txt",
nrisk_inp = "digitization_txt/CourbSurv1_nrisk.txt",
km_output = "digitization_txt/CourbSurv1_KMdata.txt",
ipd_output = "digitization_txt/CourbSurv1_IPDdata.txt")
digitise(surv_inp = "digitization_txt/CourbSurv2_time.txt",
nrisk_inp = "digitization_txt/CourbSurv2_nrisk.txt",
km_output = "digitization_txt/CourbSurv2_KMdata.txt",
ipd_output = "digitization_txt/CourbSurv2_IPDdata.txt")
data_ipd <- rbind(
read.table("digitization_txt/CourbSurv1_IPDdata.txt", header = TRUE, row.names = NULL) %>% mutate(arm = 1),
read.table("digitization_txt/CourbSurv2_IPDdata.txt", header = TRUE, row.names = NULL) %>% mutate(arm = 2)
)
# Plot
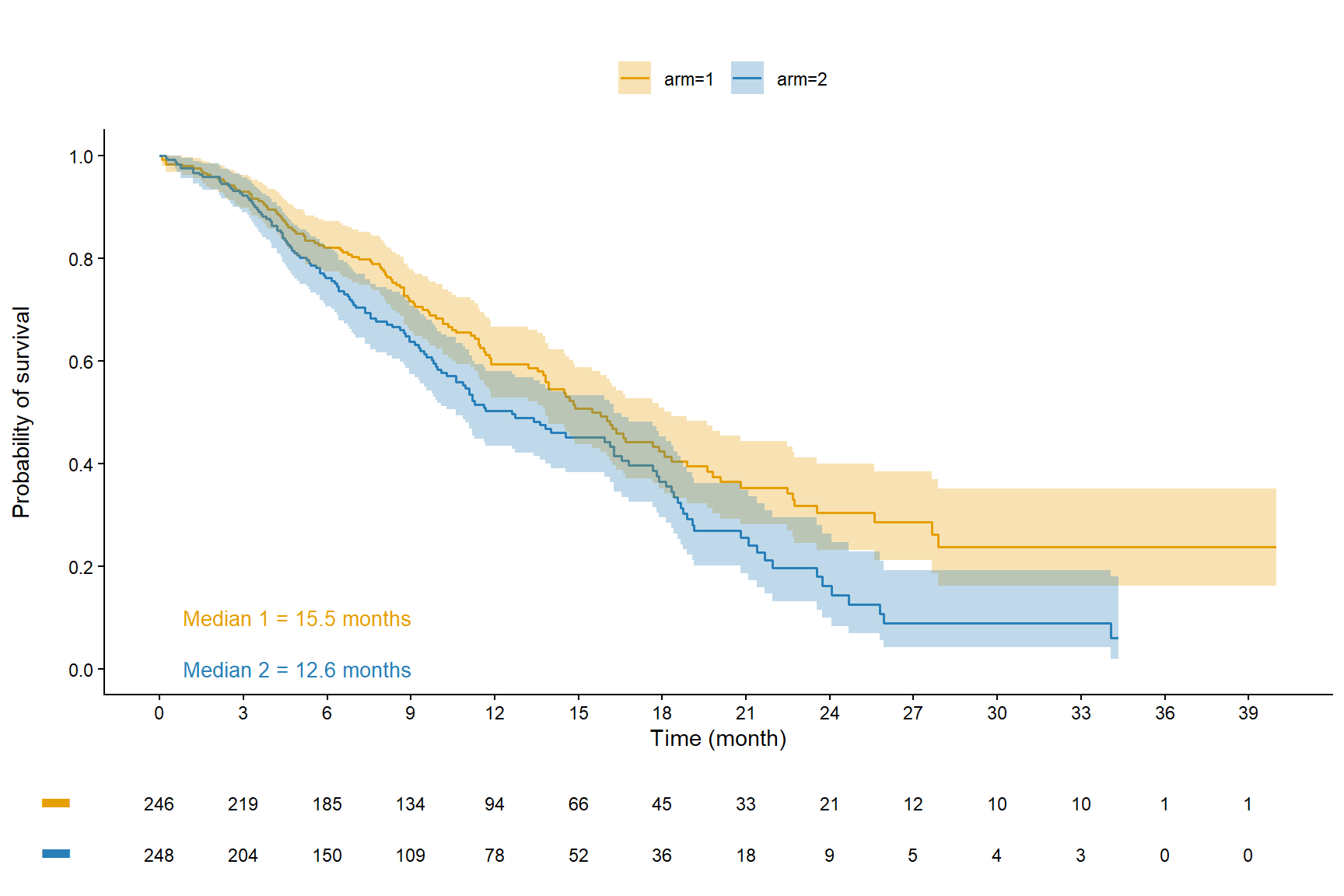
fit <- survfit(Surv(time, event)~arm, data=data_ipd)
plot.CourbSurv <- ggsurvplot(fit, conf.int=TRUE,
xlab="Time (month)",
ylab="Probability of survival",
break.time.by=3, break.y.by = 0.2,
lwd=0.6, title="",
legend="top", legend.title="",
#legend.labs=c("Arm 1","Arm 2"),
palette=c("#E69F00","#2980B9"),
censor = FALSE,
ggtheme=theme_classic(),
risk.table=TRUE,
risk.table.title="Number at risk",
risk.table.y.text=FALSE,
risk.table.fontsize=3,
tables.theme=theme_cleantable())
plot.CourbSurv$plot <- plot.CourbSurv$plot +
annotate("text", x=0, y=0.1, hjust=-0.1, size=3.5, color="#E69F00",
label=paste0("Median 1 = ", round(surv_median(fit)$median,1)[1], " months")) +
annotate("text", x=0, y=0, hjust=-0.1, size=3.5, color="#2980B9",
label=paste0("Median 2 = ", round(surv_median(fit)$median,1)[2], " months"))
plot.CourbSurv$table <- plot.CourbSurv$table + theme(plot.title = element_blank())ggarrange(plot.CourbSurv$plot, plot.CourbSurv$table, ncol = 1, nrow = 2, heights = c(0.85, 0.15), align = "v")