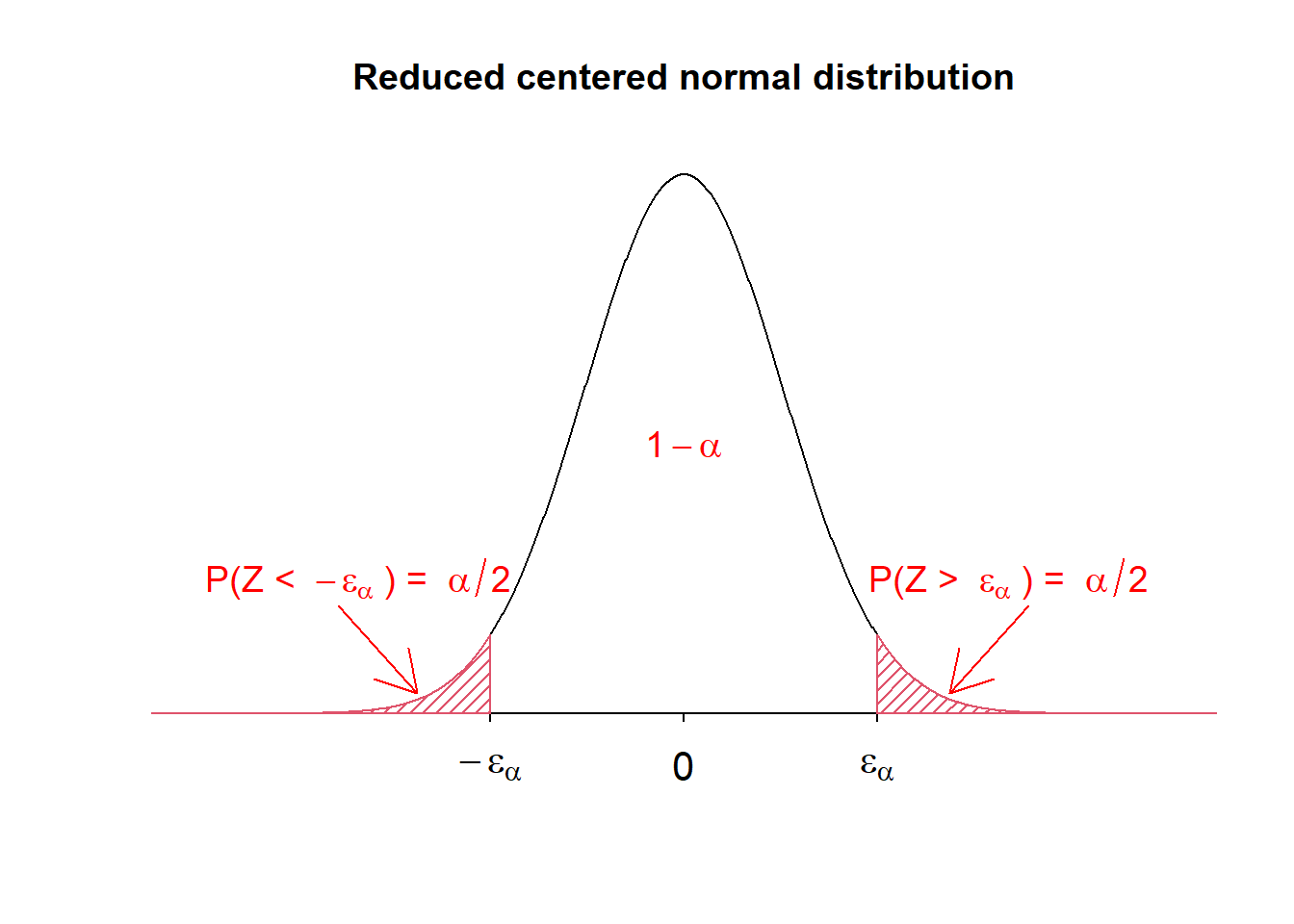
Basic density curve
A density plot is a representation of the distribution of a numeric variable. It is a smoothed version of the histogram and is used in the same kind of situation. Here is a basic example of a density plot:
# Libraries
library(ggplot2)
# Creation of dataset
set.seed(14012021)
x <- rnorm(200, mean=4)
df <- data.frame(x)
# Density curve
ggplot(df, aes(x=x)) +
geom_density() +
theme_bw()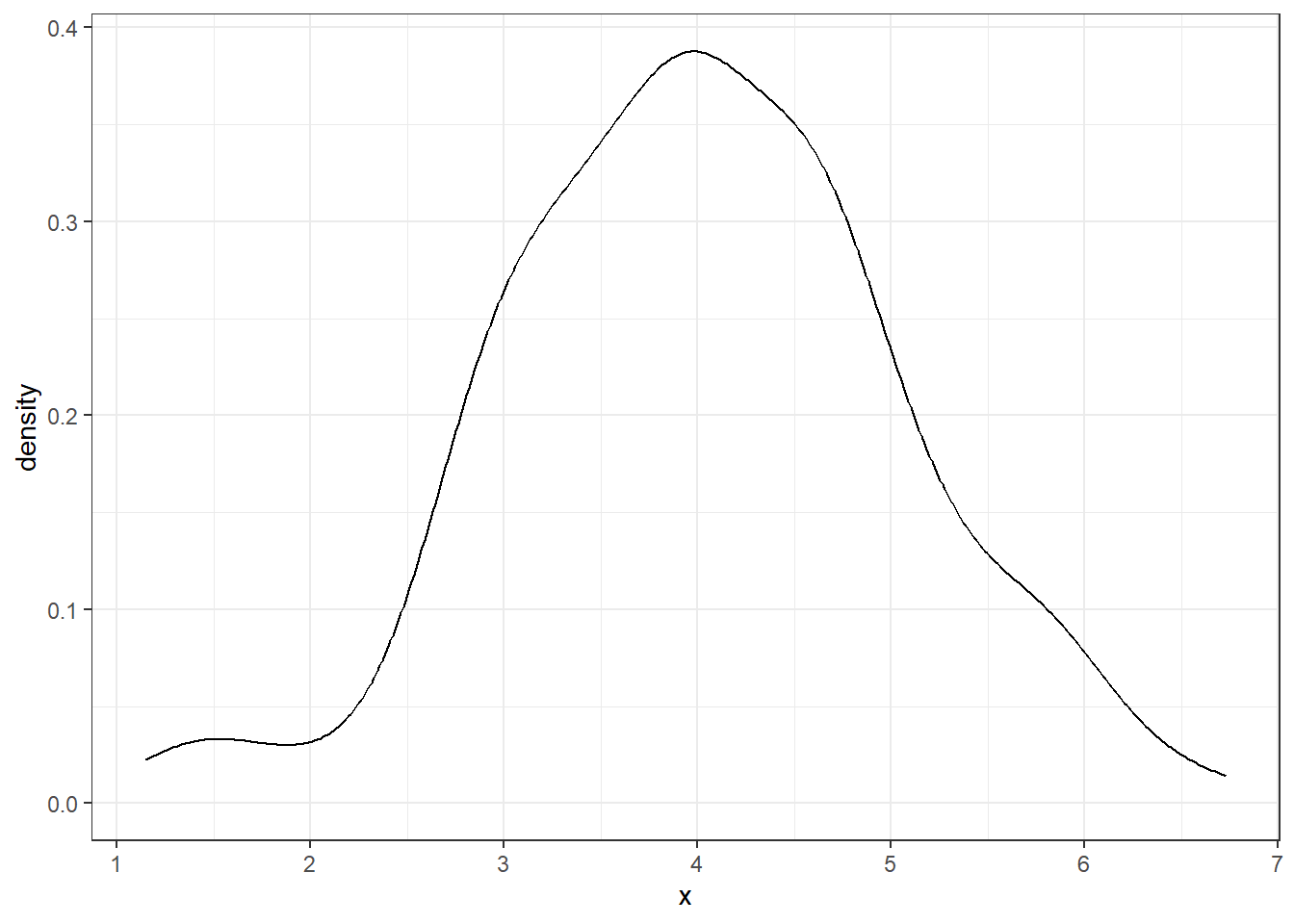
Here is a basic example of a colored density plot with a mean line:
ggplot(df, aes(x=x)) +
geom_density(color=4, fill=4, alpha=0.25) +
theme_bw() +
geom_vline(aes(xintercept = mean(x)),
color = "blue", linetype="dashed", size=1.5)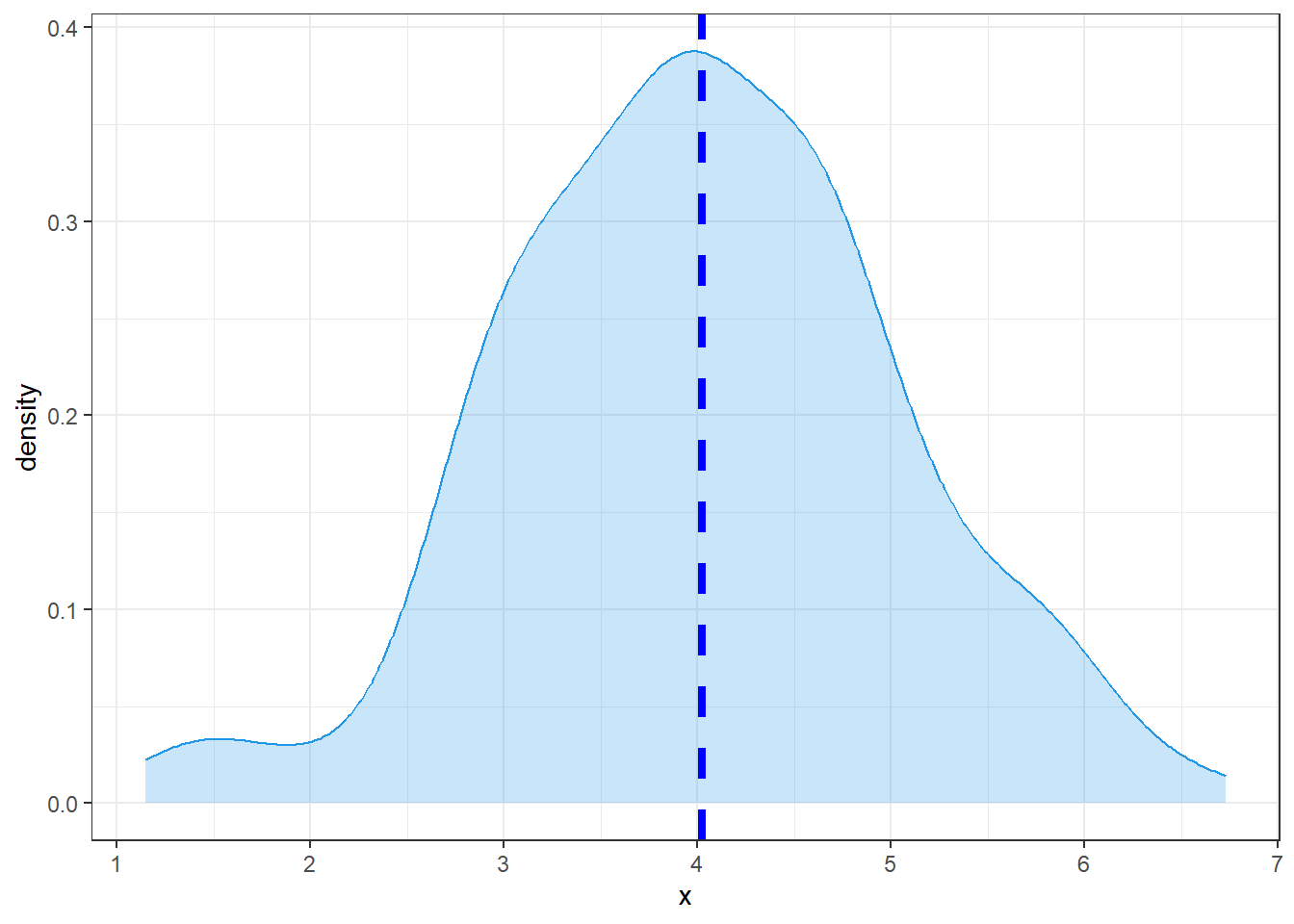
Density curve with filled area
Two-tailed filled area:
# Libraries
library(data.table)
library(ggplot2)
# Make a distribution
normalDistribution <- data.frame(
x = seq(-4,4, by = 0.01),
y = dnorm(seq(-4,4, by = 0.01))
)
criticalValues <- qnorm(c(.025,.975))
shadeNormalTwoTailedLeft <- rbind(c(criticalValues[1],0), subset(normalDistribution, x < criticalValues[1]))
shadeNormalTwoTailedRight <- rbind(c(criticalValues[2],0), subset(normalDistribution, x > criticalValues[2]), c(3,0))
ggplot(normalDistribution, aes(x,y)) +
geom_line() +
geom_polygon(data = shadeNormalTwoTailedLeft, aes(x=x, y=y, fill="red")) +
geom_polygon(data = shadeNormalTwoTailedRight, aes(x=x, y=y, fill="red")) +
geom_hline(yintercept = 0) +
theme_classic() +
geom_segment(aes(x = criticalValues[1], y = 0, xend = criticalValues[1], yend = dnorm(criticalValues[1]))) +
geom_segment(aes(x = criticalValues[2], y = 0, xend = criticalValues[2], yend = dnorm(criticalValues[2]))) 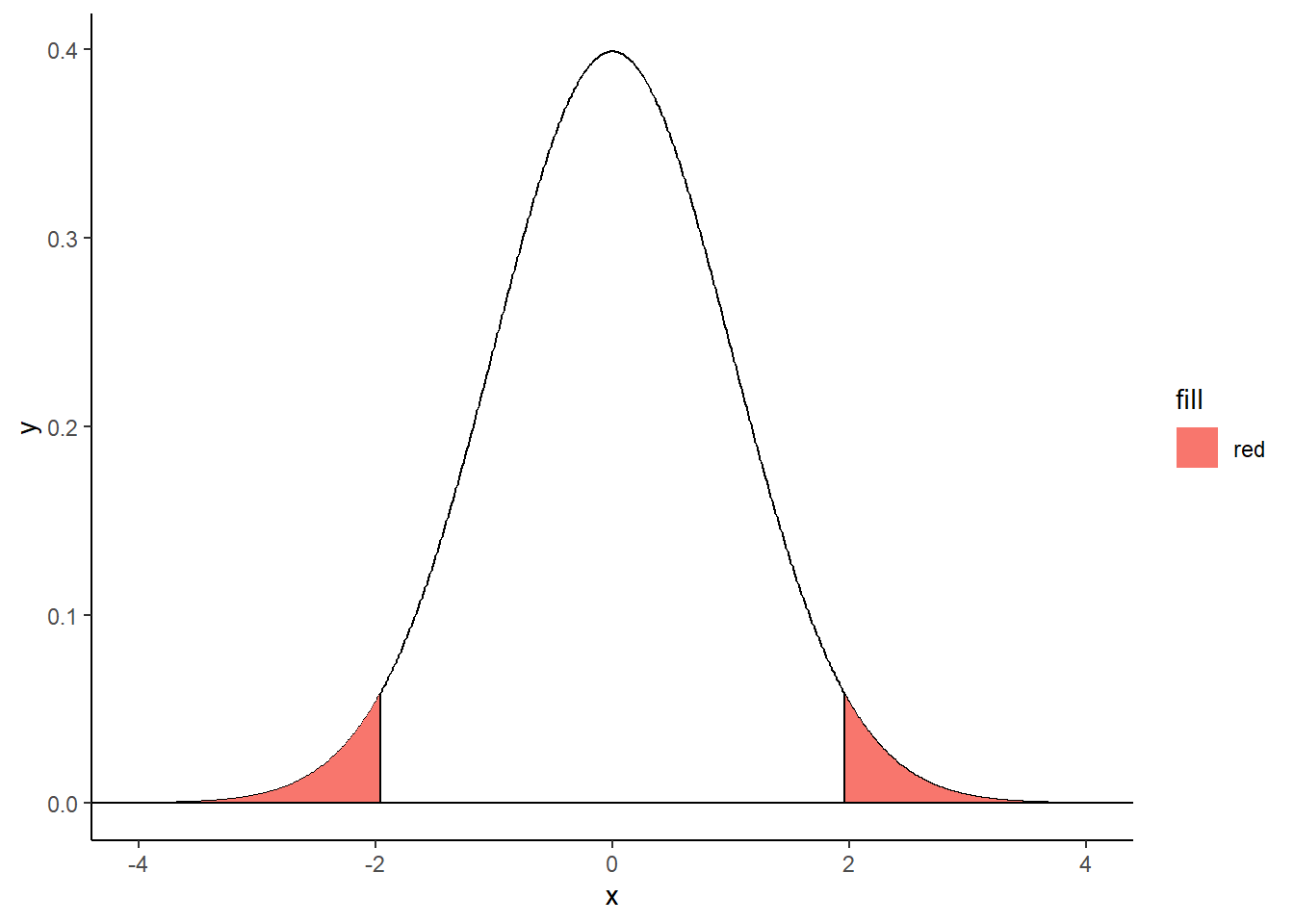
Right-tailed filled area:
criticalValue <- qnorm(.95)
shadeNormal <- rbind(c(criticalValue,0), subset(normalDistribution, x > criticalValue), c(3,0))
ggplot(normalDistribution, aes(x,y)) +
geom_line() +
geom_polygon(data = shadeNormal, aes(x=x, y=y, fill="red")) +
geom_hline(yintercept = 0) +
theme_classic() +
geom_segment(aes(x = criticalValue, y = 0, xend = criticalValue, yend = dnorm(criticalValue))) ## Warning in geom_segment(aes(x = criticalValue, y = 0, xend = criticalValue, : All aesthetics have length 1, but the data has 801 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.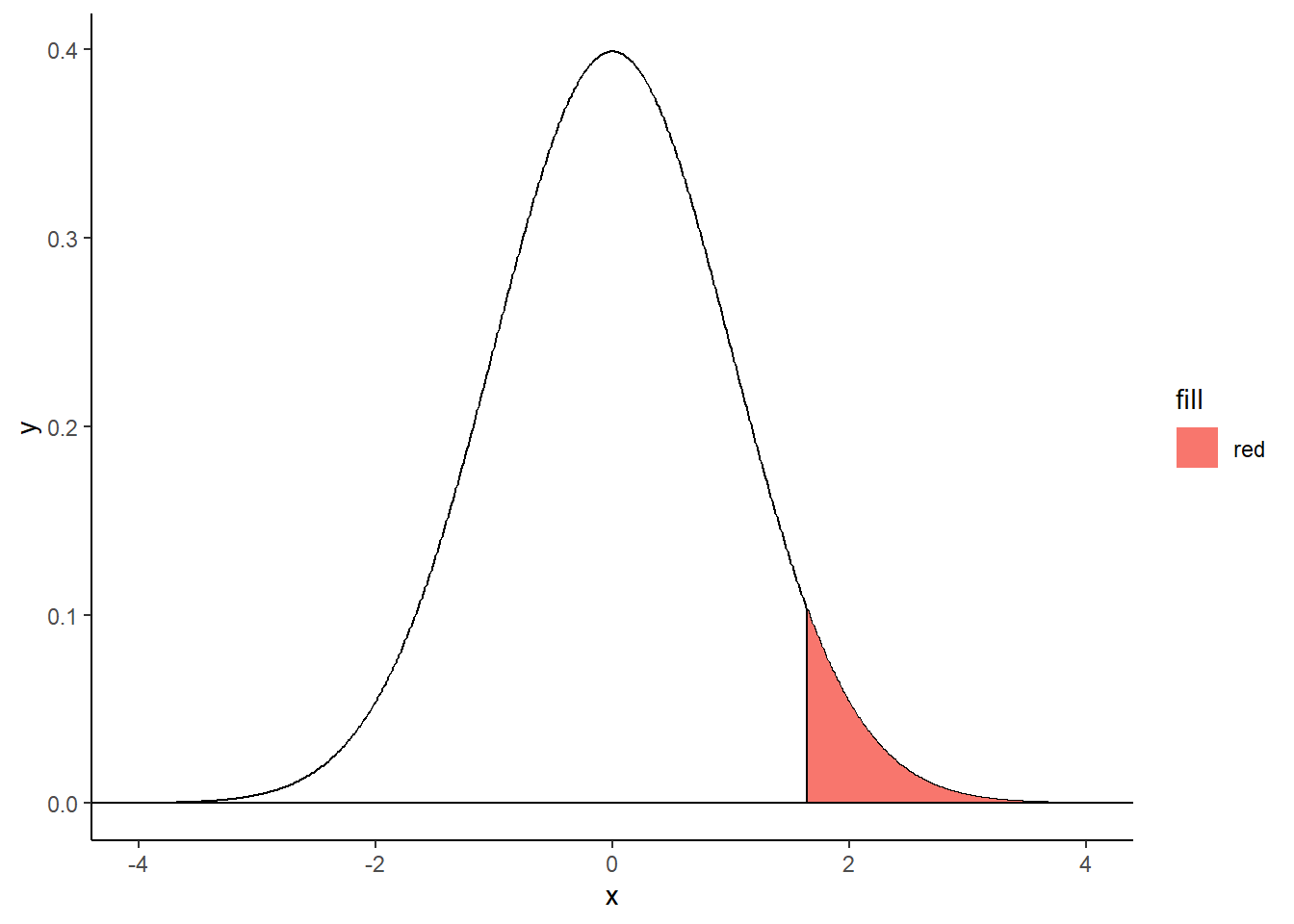
Left-tailed filled area:
criticalValue <- qnorm(.15)
shadeNormal <- rbind(c(criticalValue,0), subset(normalDistribution, x < criticalValue))
ggplot(normalDistribution, aes(x,y)) +
geom_polygon(data = shadeNormal, aes(x=x, y=y, fill="red")) +
geom_line() +
geom_hline(yintercept = 0) +
theme_classic() +
geom_segment(aes(x = criticalValue, y = 0, xend = criticalValue, yend = dnorm(criticalValue))) ## Warning in geom_segment(aes(x = criticalValue, y = 0, xend = criticalValue, : All aesthetics have length 1, but the data has 801 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.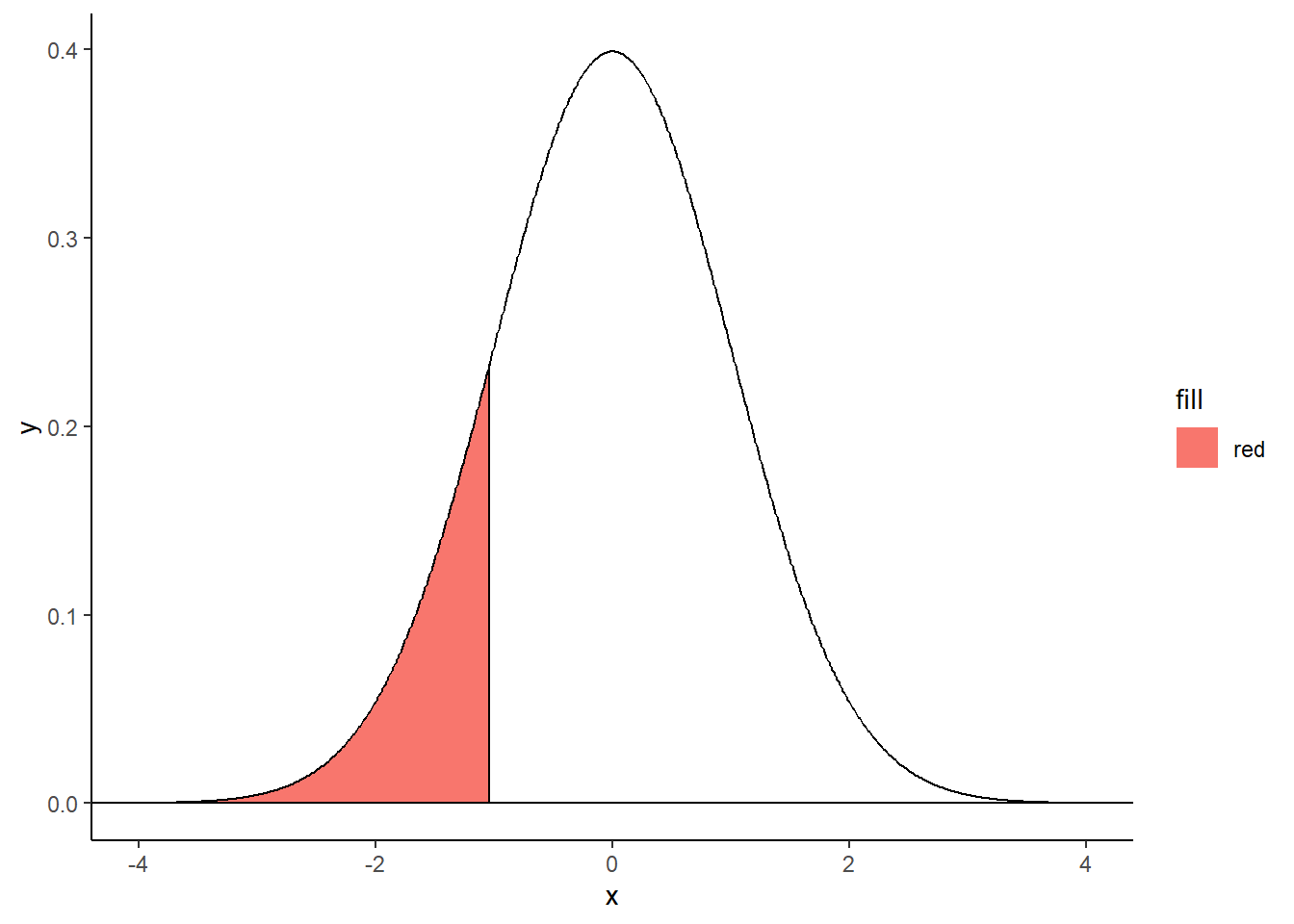
Hatched area and text:
x <- seq(-5, 5, 0.01)
plot(x, dnorm(x, 0, 1), type="l", ylim=c(0, 0.4), xlab="", ylab="",
main="Reduced centered normal distribution",
bty="n",xaxt="n", yaxt="n", lwd=1)
axis(1, pos=0, at=c(-5.5, -1.96, 0, 1.96, 5.5),
labels=c("", expression(-epsilon[alpha]), 0, expression(epsilon[alpha]), ""),
tcl=-0.2, cex.axis=1.3)
xx <- seq(qnorm(0.975), 5.5,0.01)
polygon(c(xx, rev(xx)), c(rep(0, length(xx)), rev(dnorm(xx))), density=20, col=2)
xx <- seq(-5.5, qnorm(0.025), 0.01)
polygon(c(xx, rev(xx)), c(rep(0, length(xx)), rev(dnorm(xx))), density=20, col=2)
text(3.3, 0.1, expression("P(Z > " ~ epsilon[alpha] ~ ") = " ~ alpha/2), col="red", cex=1.2)
arrows(3.5,0.08,2.7,0.015,col='red',lwd=1,code=2)
text(-3.3, 0.1, expression("P(Z < " ~ -epsilon[alpha] ~ ") = " ~ alpha/2), col="red", cex=1.2)
arrows(-3.5,0.08,-2.7,0.015,col='red',lwd=1,code=2)
text(0, 0.2, expression(1 - alpha), col="red", cex=1.2)